View sequential rules (with strings) with the Visual Pattern Viewer (SPMF documentation)
Sequential rules (with strings) are a type of patterns that can be produced by different algorithms offered in SPMF.
This page explains how to visualize the sequential rules (with strings) found by an algorithm using the Visual Pattern Viewer.
How to run this example?
If you want to run this example using the graphical user interface of SPMF, follow these steps.
1) First, select an sequential rule (with strings) mining algorithm offered in SPMF. Several algorithms are offered and are described in the documentation of SPMF.
2) Then, in the user interface of SPMF, after selecting an algorithm and setting its input file path, output file path, and parameters, click on the combo-box besides "Open output file using:", and select "Visualize_Sequential_rules_with_strings" so that the discovered patterns will be opened with the visual pattern viewer.
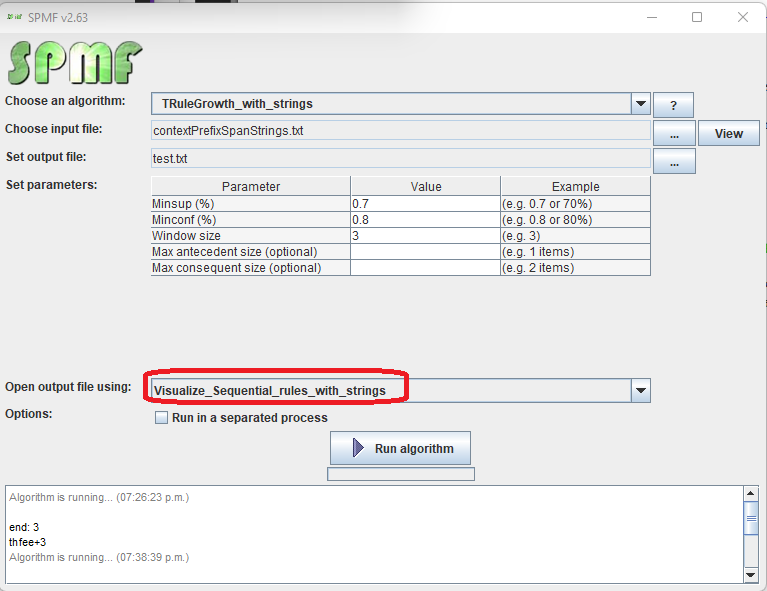
3) Then click on "Run algorithm" to run the algorithm.
After the algorithm terminates, the discovered patterns will be displayed using the Visual Pattern Viewer:
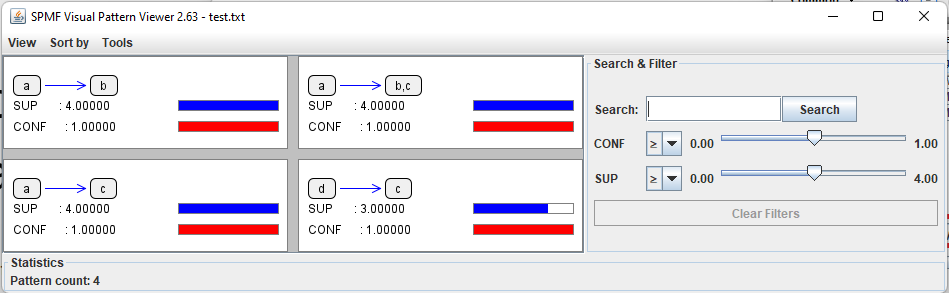
The Visual Pattern Viewer interface is quite intuitive. It displays each pattern with its value for each evaluation measure using a colored bar.
The Visual Pattern Viewer offers several features such as:
- Viewing patterns using different layouts (grid, horizontal and vertical layout).
- Sorting patterns by size and measure values.
- Searching and filtering using measure values
- Displaying statistics about the number of patterns found.
Other ways of running the Visual Pattern Viewer
It is also possible to run the Visual Pattern Viewer as an algorithm from the GUI of SPMF..
In this case, in the user interface of SPMF, select "Visualize_Sequential_rules_with_strings" as algorithm. Then, select a file containing sequential rules(with strings) as input file. Then, click "run algorithm".
This will display the patterns from the file using the Visual Pattern Viewer.
Besides, it is also possible to call the Visual Pattern Viewer from the command line interface of SPMF using this syntax:
java -jar spmf.jar run ALGORITHM_NAME PATTERN_FILE.TXT in a folder containing spmf.jar and an input file containing a pattern file, here called: PATTERN_FILE.txt.
What is the input file format?
The algorithm takes as input a file containing sequential rules (with strings) .
The file format is defined as follows. It is a text file, where each line represents an sequential rule (with strings) .
On each line, the items from the rule antecedent are first listed, separated by single spaces. Then the keyword "==>" appears, followed by the items from the rule consequent, separated by single spaces. Then, the keyword "#SUP:" appears followed by an integer indicating the support of the rule as a number of sequences. Then, the keyword "#CONF:" appears followed by a double values in the [0, 1] interval indicating the confidence of the rule.
For example, here are a few lines from a pattern file:
a ==> b #SUP: 4 #CONF: 1.0
a ==> b,c #SUP: 4 #CONF: 1.0
a ==> c #SUP: 4 #CONF: 1.0
d ==> c #SUP: 3 #CONF: 1.0
Consider the second line. It indicates that the rule {a} ==> {b, c} has a support of 4 sequences and a confidence of 100 %. The other lines follow the same format.